7 Hodgkin-Huxley neuron model
7.1 neuron_func_1
def neuron_func_1():
'''
NOTE: dvdt = I - g * (v - E)
- has stable eq at E
- looks reasonable for sub threshold dynamics
- nothing like an action potential
'''
tau = 0.01
T = 100
t = np.arange(0, T, tau)
n = t.shape[0]
I = np.zeros(n)
v = np.zeros(n)
vr = -65.0
E = -65.0
g = 1.0
C = 1.0
v[0] = vr
I[n // 5:2 * n // 5] = 30.0
I[3 * n // 5:4 * n // 5] = -30.0
for i in range(1, n):
delta_t = t[i] - t[i - 1]
dvdt = (I[i - 1] - g * (v[i - 1] - E)) / C
v[i] = v[i - 1] + dvdt * delta_t
fig, ax = plt.subplots(3, 1, squeeze=False)
ax[0, 0].plot(t, I, label='I')
ax[1, 0].plot(t, g * (v - E), label='g * (v - E)')
ax[2, 0].plot(t, v, label='v')
[x.legend() for x in ax.flatten()]
plt.tight_layout()
plt.show()
neuron_func_1()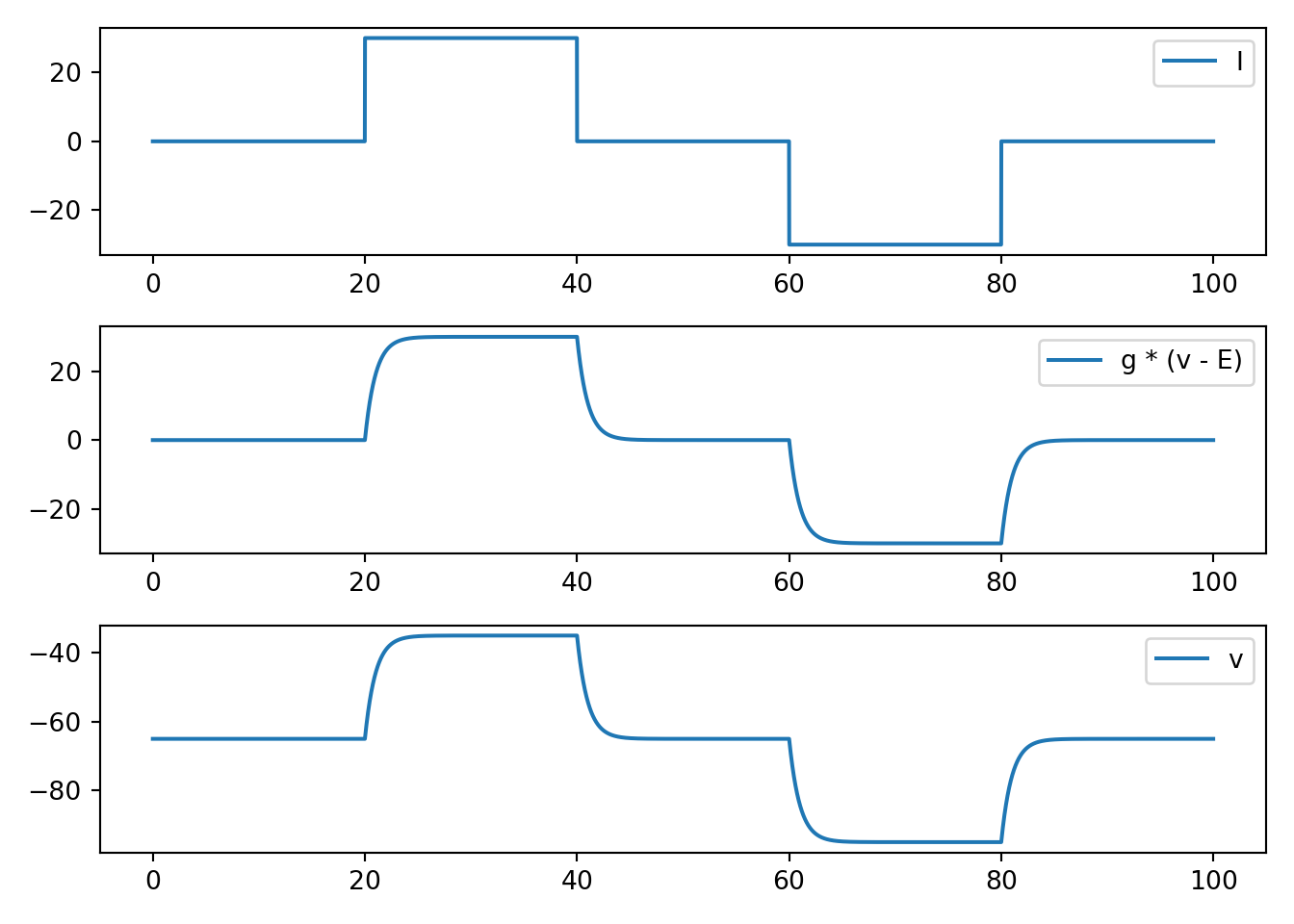
7.2 neuron_func_2
def neuron_func_2():
'''
NOTE: dvdt = I - g_a * (v - E_a) - g_b * (v - E_b)
- has stable eq between E_a and E_b with balance determined by relative g's
- still looks reasonable for sub threshold
- still nothing like an action potential
'''
tau = 0.01
T = 100
t = np.arange(0, T, tau)
n = t.shape[0]
I = np.zeros(n)
v = np.zeros(n)
vr = -65.0
E_a = -65.0
E_b = 65.0
g_a = 1.0
g_b = 1.0
C = 1.0
v[0] = vr
I[n // 5:2 * n // 5] = 30.0
I[3 * n // 5:4 * n // 5] = -30.0
for i in range(1, n):
delta_t = t[i] - t[i - 1]
dvdt = (I[i - 1] - g_a * (v[i - 1] - E_a) - g_b * (v[i - 1] - E_b)) / C
v[i] = v[i - 1] + dvdt * delta_t
fig, ax = plt.subplots(3, 1, squeeze=False)
ax[0, 0].plot(t, I, label='I')
ax[1, 0].plot(t, g_a * (v - E_a), label='g * (v - E)')
ax[1, 0].plot(t, g_b * (v - E_b), label='g * (v - E)')
ax[2, 0].plot(t, v, label='v')
[x.legend() for x in ax.flatten()]
plt.tight_layout()
plt.show()
neuron_func_2()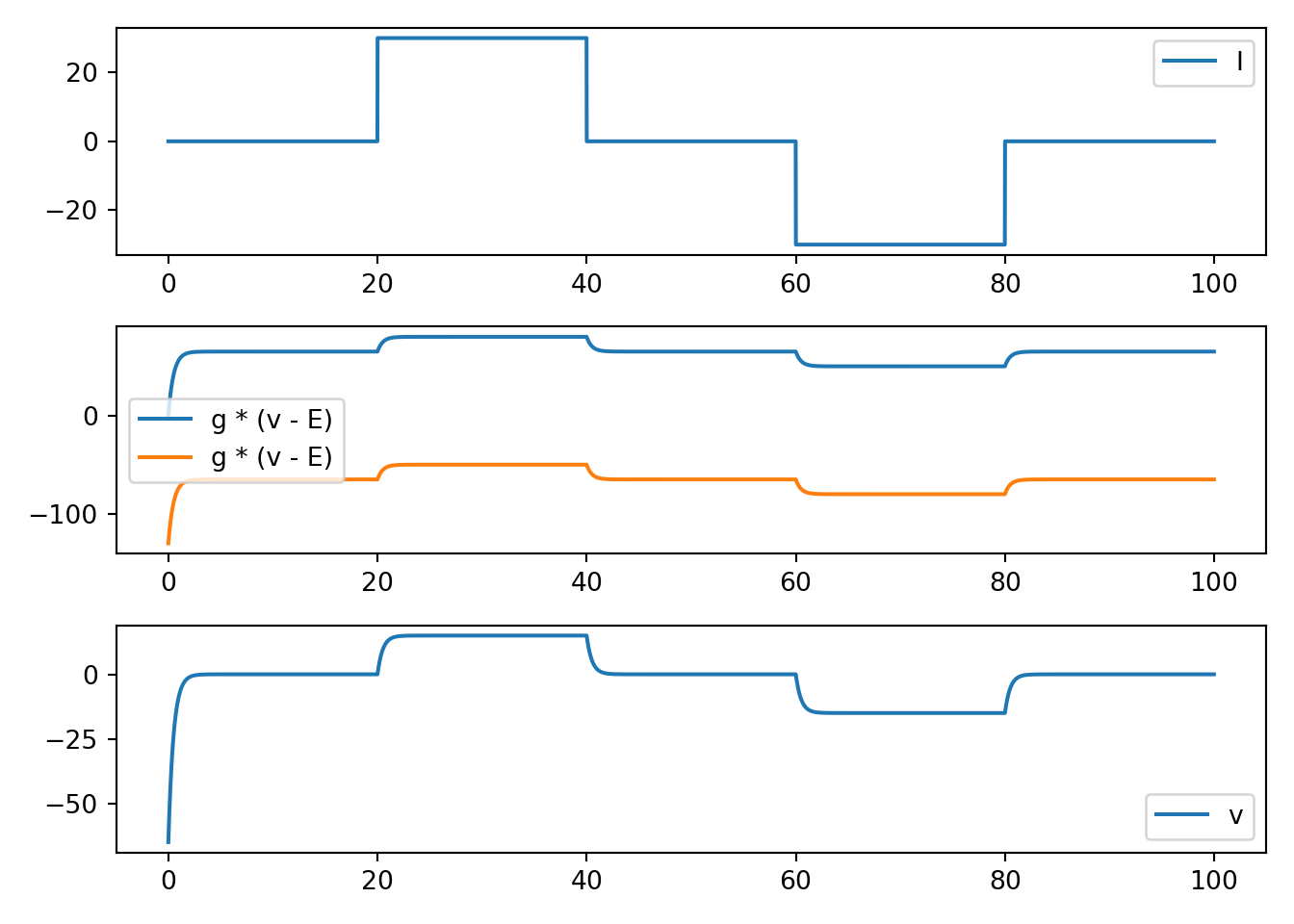
7.3 neuron_func_3
def neuron_func_3():
'''
dvdt = I - g_a_max * a * (v - E_a) - g_b * (v - E_b)
dadt = (a_inf - a) / a_tau
- Increase due to I can be amplified
- rise looks an action potential
- no fall back to rest
'''
def g_func_inf(v):
# sigmoid shape
return 1 / (1 + 0.1 * np.exp(-0.1 * (v)))
def g_func_tau(v):
# bell shape
mu = -10.0
sig = 50.0
return np.exp(-((v - mu) / sig)**2)
tau = 0.01
T = 100
t = np.arange(0, T, tau)
n = t.shape[0]
I = np.zeros(n)
v = np.zeros(n)
g_a = np.zeros(n)
g_b = np.zeros(n)
vr = -65.0
E_a = -65.0
E_b = -65.0
g_a_max = 20.0
g_b = 1.0
C = 1.0
v[0] = vr
g_a[0] = g_func_inf(vr)
I[n // 5:2 * n // 5] = 30.0
I[3 * n // 5:4 * n // 5] = -30.0
for i in range(1, n):
delta_t = t[i] - t[i - 1]
dvdt = (I[i - 1] - g_a_max * g_a[i - 1] * (v[i - 1] - E_a) - g_b *
(v[i - 1] - E_b)) / C
dgdt = (g_func_inf(v[i - 1]) - g_a[i - 1]) / g_func_tau(v[i - 1])
v[i] = v[i - 1] + dvdt * delta_t
g_a[i] = g_a[i - 1] + dgdt * delta_t
fig, ax = plt.subplots(5, 1, squeeze=False)
x = np.arange(-90, 90, 0.01)
ax[0, 0].plot(x, g_func_inf(x), label='g_func_inf')
ax[1, 0].plot(x, g_func_tau(x), label='g_func_tau')
ax[2, 0].plot(t, I, label='I')
ax[3, 0].plot(t,
g_a_max * g_a * (v - E_a),
label='g_max * g_a * (v - E_a)')
ax[3, 0].plot(t, g_b * (v - E_b), label='g_b * (v - E_b)')
ax[4, 0].plot(t, v, label='v')
[x.legend() for x in ax.flatten()]
plt.tight_layout()
plt.show()
neuron_func_3()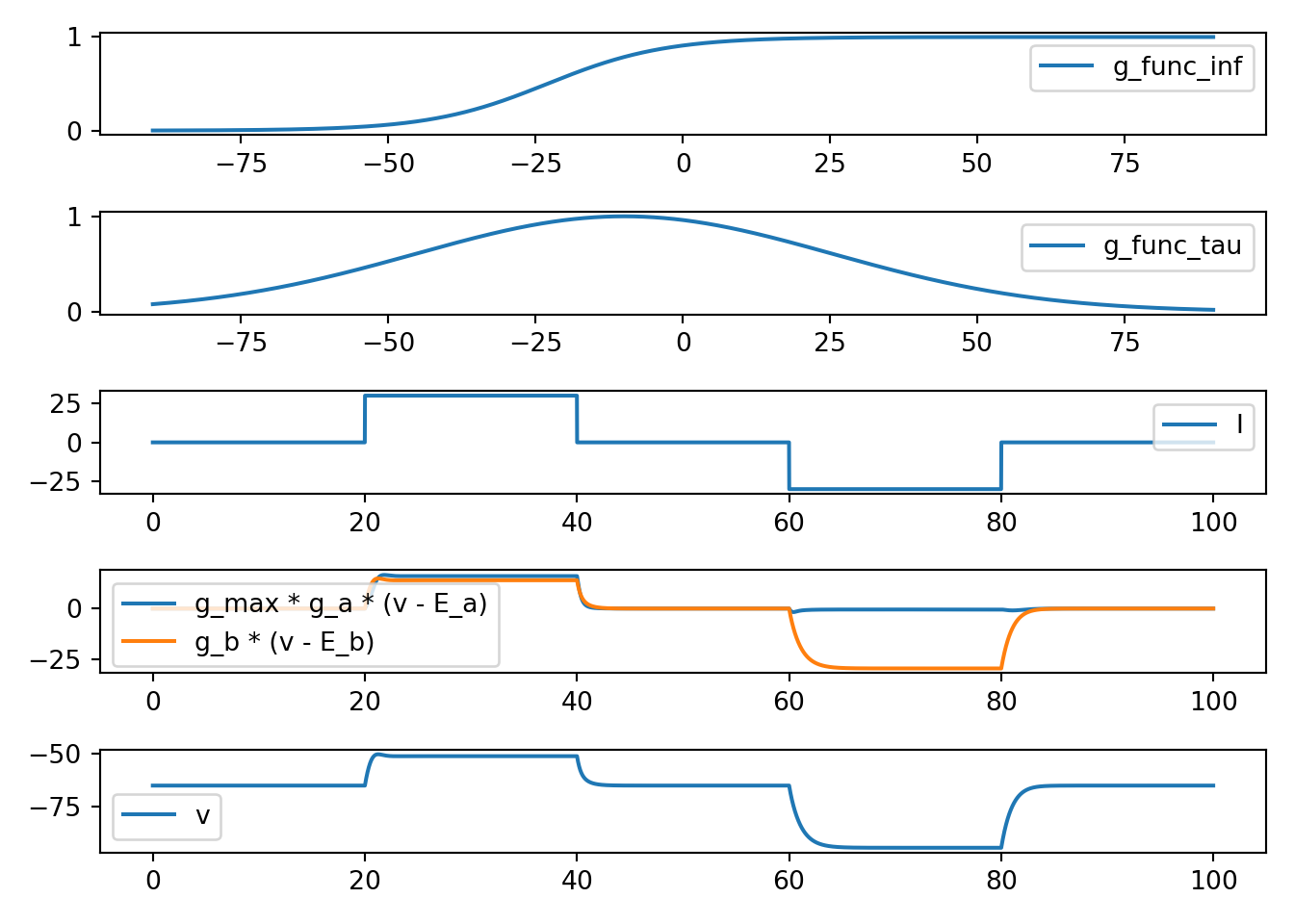
7.4 neuron_func_4
def neuron_func_4():
'''
dvdt = I - g_a_max * a * (v - E_a) - g_b_max * b * (v - E_b)
dadt = (a_inf - a) / a_tau
dbdt = (b_inf - b) / b_tau
- Increase due to I can be amplified
- rise looks an action potential (due to a)
- can get a fall back to E_b (due to b)
- fall occurs too slowly to be a normal action potential
'''
def g_func_inf_a(v):
# sigmoid shape
return 1 / (1 + 0.1 * np.exp(-0.1 * (v)))
def g_func_inf_b(v):
# sigmoid shape
return -1 / (1.1 + 0.1 * np.exp(-0.1 * (v))) + 1
def g_func_tau_a(v):
# bell shape
mu = -10.0
sig = 60.0
return (300 / sig) * np.exp(-((v - mu) / sig)**2)
def g_func_tau_b(v):
# bell shape
mu = -10.0
sig = 50.0
return (300 / sig) * np.exp(-((v - mu) / sig)**2)
tau = 0.01
T = 200
t = np.arange(0, T, tau)
n = t.shape[0]
I = np.zeros(n)
v = np.zeros(n)
g_a = np.zeros(n)
g_b = np.zeros(n)
vr = -65.0
E_a = 120.0
E_b = -70.0
g_a_max = 1.0
g_b_max = 2.0
C = 1.0
v[0] = vr
g_a[0] = g_func_inf_a(vr)
g_b[0] = g_func_inf_b(vr)
I[n // 5:2 * n // 5] = 30.0
I[3 * n // 5:4 * n // 5] = -30.0
for i in range(1, n):
delta_t = t[i] - t[i - 1]
dvdt = (I[i - 1] - g_a_max * g_a[i - 1] *
(v[i - 1] - E_a) - g_b_max * g_b[i - 1] * (v[i - 1] - E_b)) / C
dgdt_a = (g_func_inf_a(v[i - 1]) - g_a[i - 1]) / g_func_tau_a(v[i - 1])
dgdt_b = (g_func_inf_b(v[i - 1]) - g_b[i - 1]) / g_func_tau_b(v[i - 1])
v[i] = v[i - 1] + dvdt * delta_t
g_a[i] = g_a[i - 1] + dgdt_a * delta_t
g_b[i] = g_b[i - 1] + dgdt_b * delta_t
fig, ax = plt.subplots(5, 1, squeeze=False)
x = np.arange(-90, 90, 0.01)
ax[0, 0].plot(x, g_func_inf_a(x), label='g_func_inf_a')
ax[0, 0].plot(x, g_func_inf_b(x), label='g_func_inf_b')
ax[1, 0].plot(x, g_func_tau_a(x), label='g_func_tau_a')
ax[1, 0].plot(x, g_func_tau_b(x), label='g_func_tau_b')
ax[2, 0].plot(t, I, label='I')
ax[3, 0].plot(t,
g_a_max * g_a * (v - E_a),
label='g_a_max * g_a * (v - E_a)')
ax[3, 0].plot(t,
g_b_max * g_b * (v - E_b),
label='g_b_max * g_b * (v - E_b)')
ax[4, 0].plot(t, v, label='v')
[x.legend() for x in ax.flatten()]
plt.tight_layout()
plt.show()
neuron_func_4()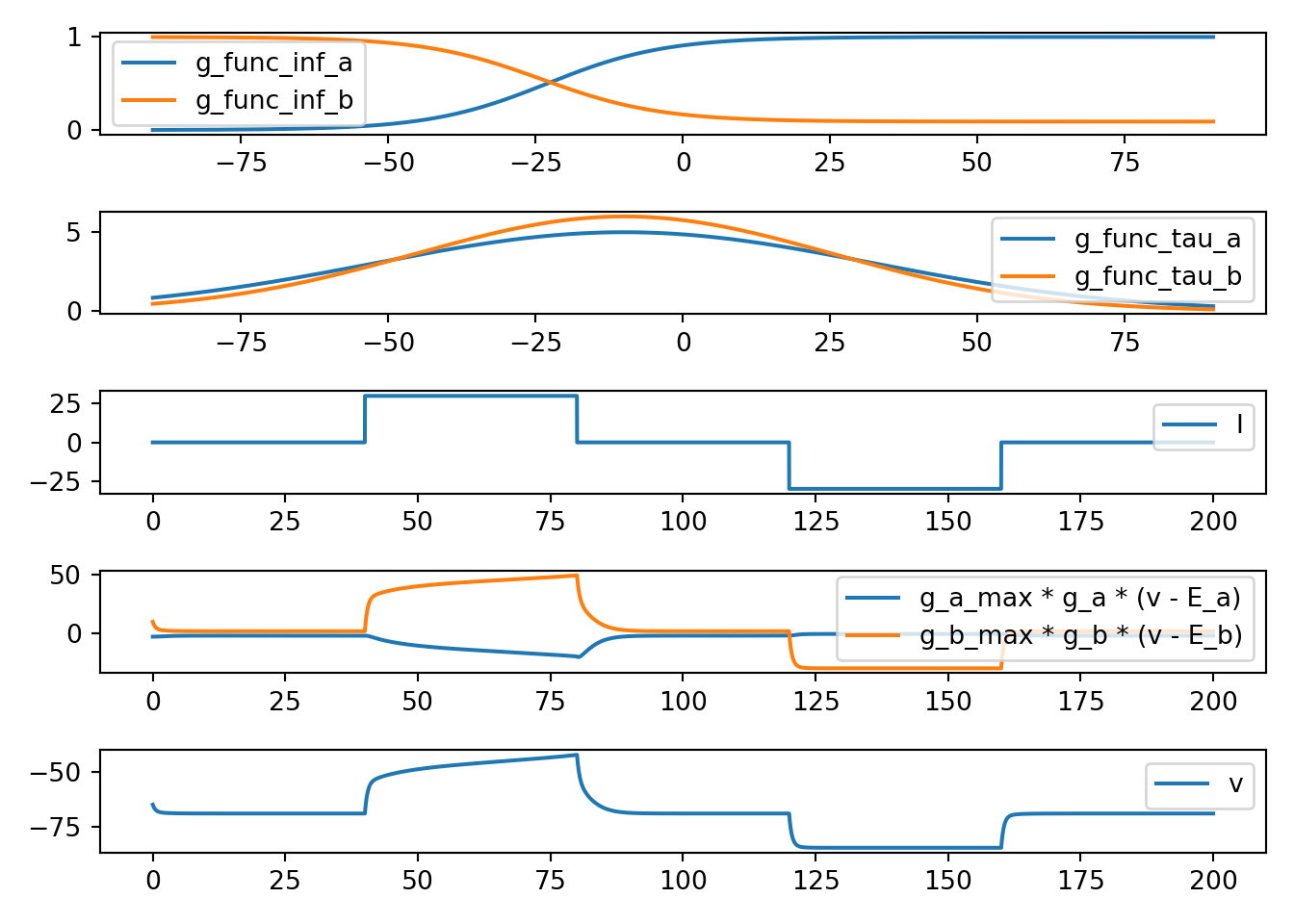
7.5 neuron_func_5
def neuron_func_5():
'''
dvdt = I - g_a_max * a_act**3 * a_inact * (v - E_a) - g_b * (v - E_b)
dadt_act = (a_act_inf - a_act) / a_act_tau
dadt_inact = (a_inact_inf - a_inact) / a_inact_tau
- Increase due to I can be amplified
- rise looks an action potential (due to a_act)
- fall back to E_b (due to a_inact)
- still not quite right... but why really?
'''
def g_func_inf_a_act(v):
# sigmoid shape
return 1 / (1 + 0.1 * np.exp(-0.5 * (v + 50.0)))
def g_func_inf_a_inact(v):
# sigmoid shape
return -1 / (1.1 + 0.1 * np.exp(-0.1 * (v))) + 1
def g_func_tau_a_act(v):
# bell shape
mu = -10.0
sig = 60.0
return (300 / sig) * np.exp(-((v - mu) / sig)**2)
def g_func_tau_a_inact(v):
# bell shape
mu = -10.0
sig = 50.0
return (300 / sig) * np.exp(-((v - mu) / sig)**2)
tau = 0.01
T = 200
t = np.arange(0, T, tau)
n = t.shape[0]
I = np.zeros(n)
v = np.zeros(n)
g_a_act = np.zeros(n)
g_a_inact = np.zeros(n)
vr = -65.0
E_a = 120.0
E_b = -70.0
g_a_max = 2.0
g_b_max = 1.0
C = 1.0
v[0] = vr
g_a_act[0] = g_func_inf_a_act(vr)
g_a_inact[0] = g_func_inf_a_inact(vr)
I[n // 5:2 * n // 5] = 30.0
I[3 * n // 5:4 * n // 5] = -30.0
for i in range(1, n):
delta_t = t[i] - t[i - 1]
dvdt = (I[i - 1] - g_a_max * g_a_act[i - 1]**3 * g_a_inact[i - 1] *
(v[i - 1] - E_a) - g_b_max * (v[i - 1] - E_b)) / C
dgdt_a_act = (g_func_inf_a_act(v[i - 1]) -
g_a_act[i - 1]) / g_func_tau_a_act(v[i - 1])
dgdt_a_inact = (g_func_inf_a_inact(v[i - 1]) -
g_a_inact[i - 1]) / g_func_tau_a_inact(v[i - 1])
v[i] = v[i - 1] + dvdt * delta_t
g_a_act[i] = g_a_act[i - 1] + dgdt_a_act * delta_t
g_a_inact[i] = g_a_inact[i - 1] + dgdt_a_inact * delta_t
fig, ax = plt.subplots(5, 1, squeeze=False)
x = np.arange(-90, 90, 0.01)
ax[0, 0].plot(x, g_func_inf_a_act(x), label='g_func_inf_a_act')
ax[0, 0].plot(x, g_func_inf_a_inact(x), label='g_func_inf_a_inact')
ax[1, 0].plot(x, g_func_tau_a_act(x), label='g_func_tau_a_act')
ax[1, 0].plot(x, g_func_tau_a_inact(x), label='g_func_tau_a_inact')
ax[2, 0].plot(t, I, label='I')
ax[3, 0].plot(t,
g_a_max * g_a_act * (v - E_a),
label='g_a_max * g_a_act * (v - E_a)')
ax[3, 0].plot(t,
g_a_max * g_a_inact * (v - E_a),
label='g_a_max * g_a_inact * (v - E_a)')
ax[4, 0].plot(t, v, label='v')
[x.legend() for x in ax.flatten()]
plt.tight_layout()
plt.show()
neuron_func_5()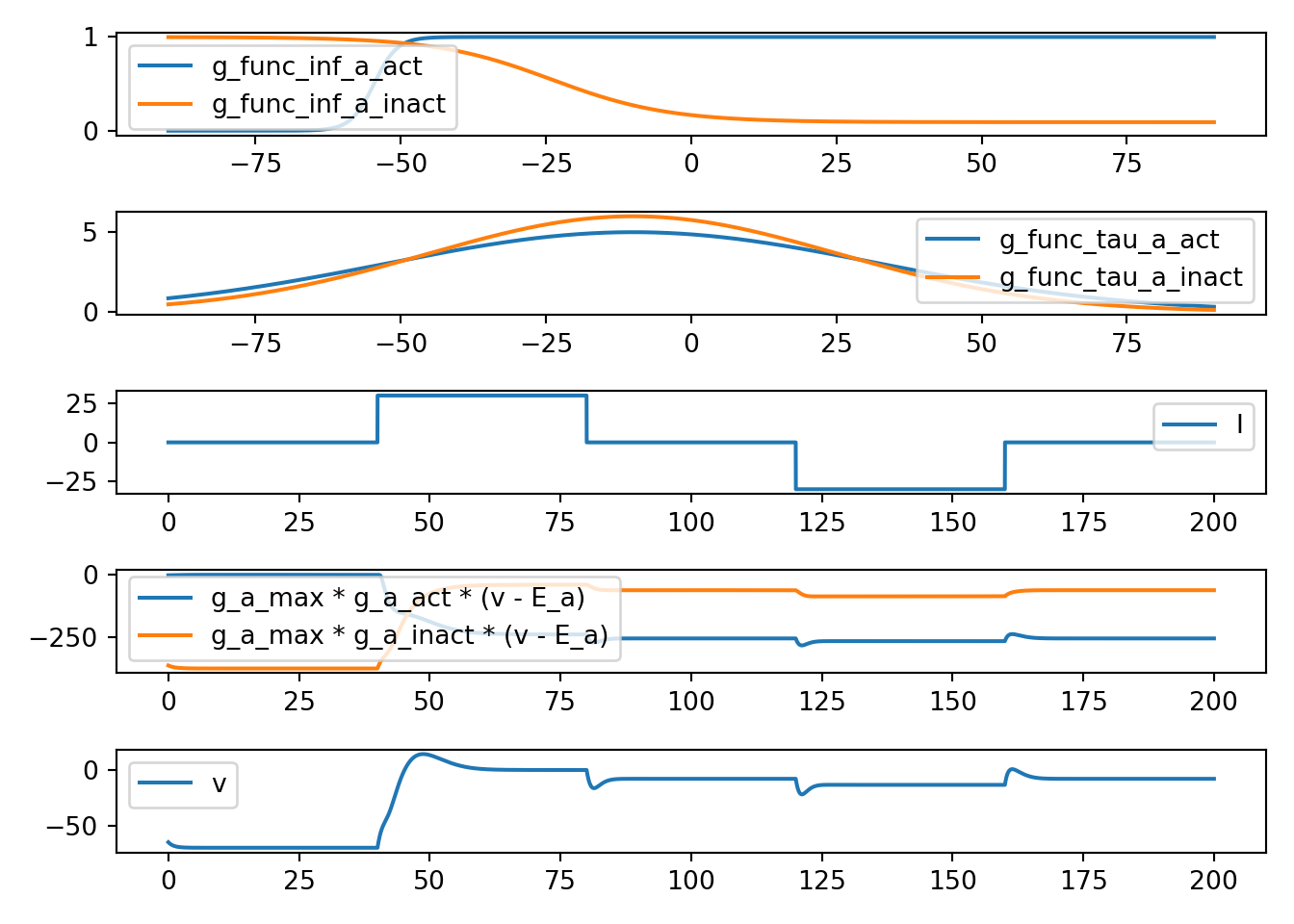
7.6 neuron_func_6
def neuron_func_6():
'''
dvdt = I - g_a_max * a_act**3 * a_inact * (v - E_a) - g_b_max * b * (v - E_b)
dadt_act = (a_act_inf - a_act) / a_act_tau
dadt_inact = (a_inact_inf - a_inact) / a_inact_tau
dbdt = (b_inf - b) / b_tau
- Increase due to I can be amplified
- rise looks an action potential (due to a_act)
- fall back to E_b (due to a_inact and due to b)
- looking prettty good presumably
'''
def g_func_inf_a_act(v):
# sigmoid shape
return 1 / (1 + 0.1 * np.exp(-0.5 * (v + 50.0)))
def g_func_inf_a_inact(v):
# sigmoid shape
return -1 / (1.1 + 0.1 * np.exp(-0.1 * (v))) + 1
def g_func_inf_b(v):
# sigmoid shape
return 1 / (1.1 + 0.1 * np.exp(-0.09 * (v)))
def g_func_tau_a_act(v):
# bell shape
mu = -10.0
sig = 60.0
return (300 / sig) * np.exp(-((v - mu) / sig)**2)
def g_func_tau_a_inact(v):
# bell shape
mu = -10.0
sig = 50.0
return (300 / sig) * np.exp(-((v - mu) / sig)**2)
def g_func_tau_b(v):
# bell shape
mu = 5.0
sig = 50.0
return (300 / sig) * np.exp(-((v - mu) / sig)**2)
tau = 0.01
T = 200
t = np.arange(0, T, tau)
n = t.shape[0]
I = np.zeros(n)
v = np.zeros(n)
g_a_act = np.zeros(n)
g_a_inact = np.zeros(n)
g_b = np.zeros(n)
vr = -65.0
E_a = 120.0
E_b = -70.0
g_a_max = 2.0
g_b_max = 5.0
C = 1.0
v[0] = vr
g_a_act[0] = g_func_inf_a_act(vr)
g_a_inact[0] = g_func_inf_a_inact(vr)
g_b[0] = g_func_inf_b(vr)
I[n // 5:2 * n // 5] = 30.0
I[3 * n // 5:4 * n // 5] = -30.0
for i in range(1, n):
delta_t = t[i] - t[i - 1]
dvdt = (I[i - 1] - g_a_max * g_a_act[i - 1]**3 * g_a_inact[i - 1] *
(v[i - 1] - E_a) - g_b_max * g_b[i - 1]**4 *
(v[i - 1] - E_b)) / C
dgdt_a_act = (g_func_inf_a_act(v[i - 1]) -
g_a_act[i - 1]) / g_func_tau_a_act(v[i - 1])
dgdt_a_inact = (g_func_inf_a_inact(v[i - 1]) -
g_a_inact[i - 1]) / g_func_tau_a_inact(v[i - 1])
dgdt_b = (g_func_inf_b(v[i - 1]) - g_b[i - 1]) / g_func_tau_b(v[i - 1])
v[i] = v[i - 1] + dvdt * delta_t
g_a_act[i] = g_a_act[i - 1] + dgdt_a_act * delta_t
g_a_inact[i] = g_a_inact[i - 1] + dgdt_a_inact * delta_t
g_b[i] = g_b[i - 1] + dgdt_b * delta_t
fig, ax = plt.subplots(5, 1, squeeze=False)
x = np.arange(-90, 90, 0.01)
ax[0, 0].plot(x, g_func_inf_a_act(x), label='g_func_inf_a_act')
ax[0, 0].plot(x, g_func_inf_a_inact(x), label='g_func_inf_a_inact')
ax[0, 0].plot(x, g_func_inf_b(x), label='g_func_inf_b')
ax[1, 0].plot(x, g_func_tau_a_act(x), label='g_func_tau_a_act')
ax[1, 0].plot(x, g_func_tau_a_inact(x), label='g_func_tau_a_inact')
ax[1, 0].plot(x, g_func_tau_b(x), label='g_func_tau_b')
ax[2, 0].plot(t, I, label='I')
ax[3, 0].plot(t,
g_a_max * g_a_act * (v - E_a),
label='g_a_max * g_a_act * (v - E_a)')
ax[3, 0].plot(t,
g_a_max * g_a_inact * (v - E_a),
label='g_a_max * g_a_inact * (v - E_a)')
ax[3, 0].plot(t,
g_b_max * g_b * (v - E_b),
label='g_b_max * g_b * (v - E_b)')
ax[4, 0].plot(t, v, label='v')
[x.legend() for x in ax.flatten()]
plt.tight_layout()
plt.show()
neuron_func_6()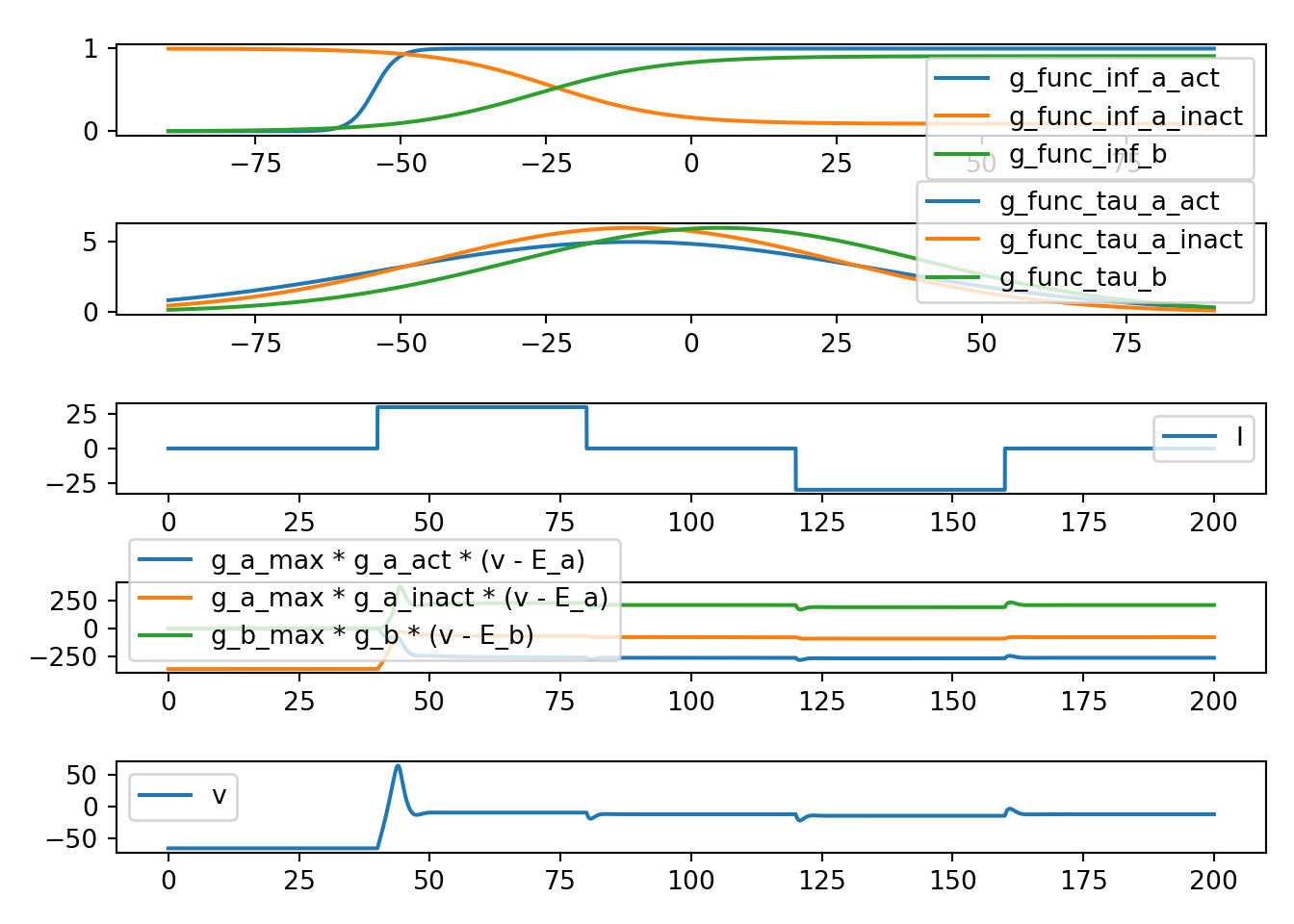
7.7 neuron_func_7
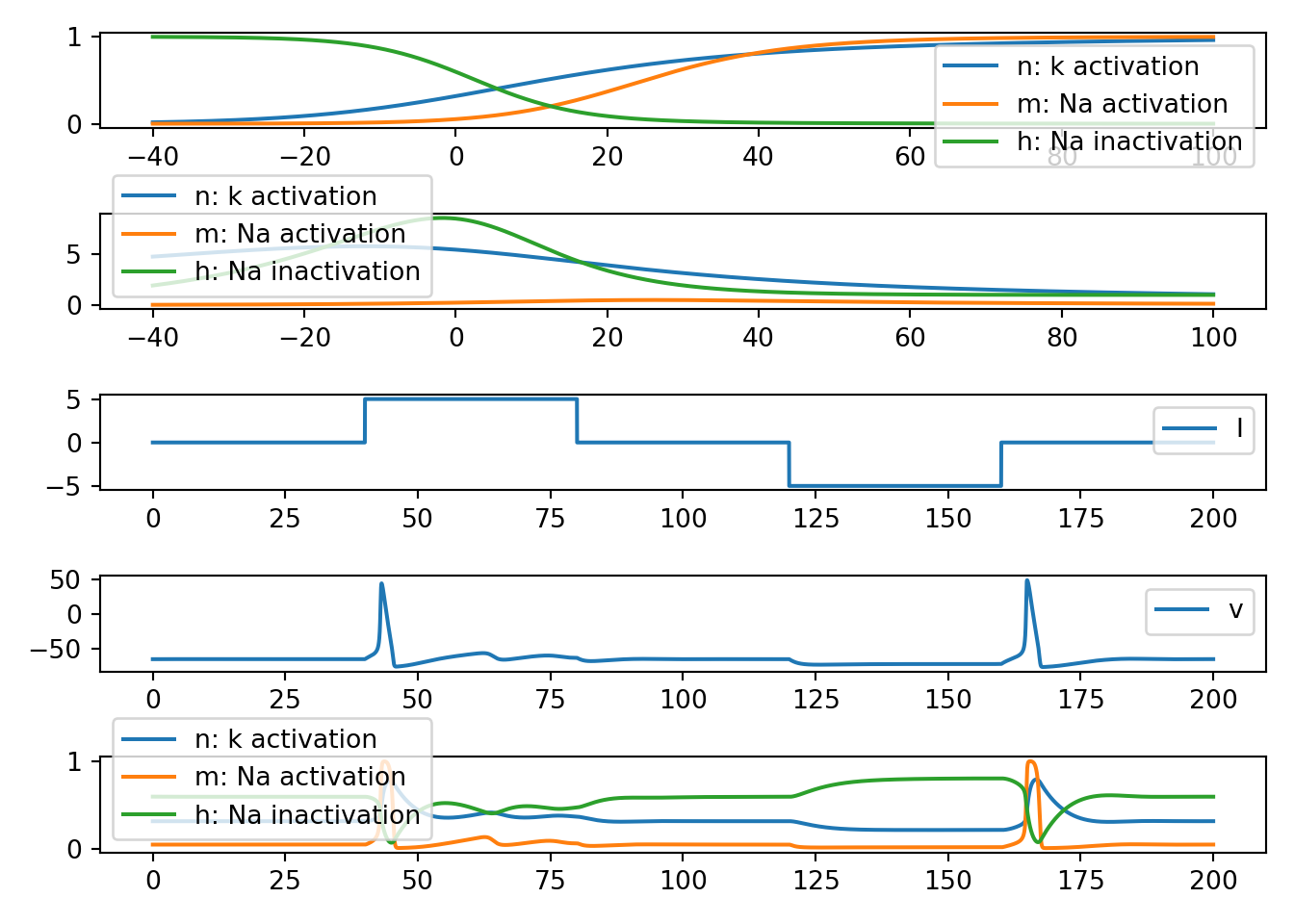
def neuron_func_7():
'''
NOTE: Full HH
'''
# NOTE: n gating variables
def n_inf(v):
return alpha_func_n(v) / (alpha_func_n(v) + beta_func_n(v))
def n_tau(v):
return 1.0 / (alpha_func_n(v) + beta_func_n(v))
def alpha_func_n(v):
return 0.01 * (10.0 - v) / (np.exp((10.0 - v) / 10.0) - 1.0)
def beta_func_n(v):
return 0.125 * np.exp(-v / 80.0)
# NOTE: m gating variables
def m_inf(v):
return alpha_func_m(v) / (alpha_func_m(v) + beta_func_m(v))
def m_tau(v):
return 1.0 / (alpha_func_m(v) + beta_func_m(v))
def alpha_func_m(v):
return 0.1 * (25.0 - v) / (np.exp((25.0 - v) / 10.0) - 1.0)
def beta_func_m(v):
return 4.0 * np.exp(-v / 18.0)
# NOTE: h gating variables
def h_inf(v):
return alpha_func_h(v) / (alpha_func_h(v) + beta_func_h(v))
def h_tau(v):
return 1.0 / (alpha_func_h(v) + beta_func_h(v))
def alpha_func_h(v):
return 0.07 * np.exp(-v / 20.0)
def beta_func_h(v):
return 1.0 / (np.exp((30.0 - v) / 10.0) + 1.0)
# NOTE: simulation parameter etc
tau = 0.01
T = 200
t = np.arange(0, T, tau)
nn = t.shape[0]
I = np.zeros(nn)
v = np.zeros(nn)
n = np.zeros(nn)
m = np.zeros(nn)
h = np.zeros(nn)
vr = -65.0
v[0] = vr
n[0] = n_inf(vr*0)
m[0] = m_inf(vr*0)
h[0] = h_inf(vr*0)
g_k = 36.0
g_na = 120.0
g_leak = 0.30
E_k = -12 + vr
E_na = 120 + vr
E_leak = 10.6 + vr
C = 1.0
I[nn // 5:2 * nn // 5] = 5.0
I[3 * nn // 5:4 * nn // 5] = -5.0
# NOTE: Euler's method simulation
for i in range(1, nn):
delta_t = t[i] - t[i - 1]
I_k = g_k * (n[i - 1]**4) * (v[i - 1] - E_k)
I_na = g_na * (m[i - 1]**3) * (h[i - 1]) * (v[i - 1] - E_na)
I_leak = g_leak * (v[i - 1] - E_leak)
dvdt = (I[i - 1] - (I_k + I_na + I_leak)) / C
dndt = (n_inf(v[i - 1] - vr) - n[i - 1]) / n_tau(v[i - 1] - vr)
dmdt = (m_inf(v[i - 1] - vr) - m[i - 1]) / m_tau(v[i - 1] - vr)
dhdt = (h_inf(v[i - 1] - vr) - h[i - 1]) / h_tau(v[i - 1] - vr)
v[i] = v[i - 1] + dvdt * delta_t
n[i] = n[i - 1] + dndt * delta_t
m[i] = m[i - 1] + dmdt * delta_t
h[i] = h[i - 1] + dhdt * delta_t
# NOTE: inspect gating functions
fig, ax = plt.subplots(5, 1, squeeze=False)
v_range = np.arange(-40, 100, 0.01)
ax[0, 0].plot(v_range, n_inf(v_range), label='n: k activation')
ax[0, 0].plot(v_range, m_inf(v_range), label='m: Na activation')
ax[0, 0].plot(v_range, h_inf(v_range), label='h: Na inactivation')
ax[1, 0].plot(v_range, n_tau(v_range), label='n: k activation')
ax[1, 0].plot(v_range, m_tau(v_range), label='m: Na activation')
ax[1, 0].plot(v_range, h_tau(v_range), label='h: Na inactivation')
ax[2, 0].plot(t, I, label='I')
ax[3, 0].plot(t, v, label='v')
ax[4, 0].plot(t, n, label='n: k activation')
ax[4, 0].plot(t, m, label='m: Na activation')
ax[4, 0].plot(t, h, label='h: Na inactivation')
[x.legend() for x in ax.flatten()]
plt.tight_layout()
plt.show()
neuron_func_7()